Zehra Camlica
Medical Image Classification via SVM using LBP Features from Saliency-Based Folded Data
Sep 15, 2015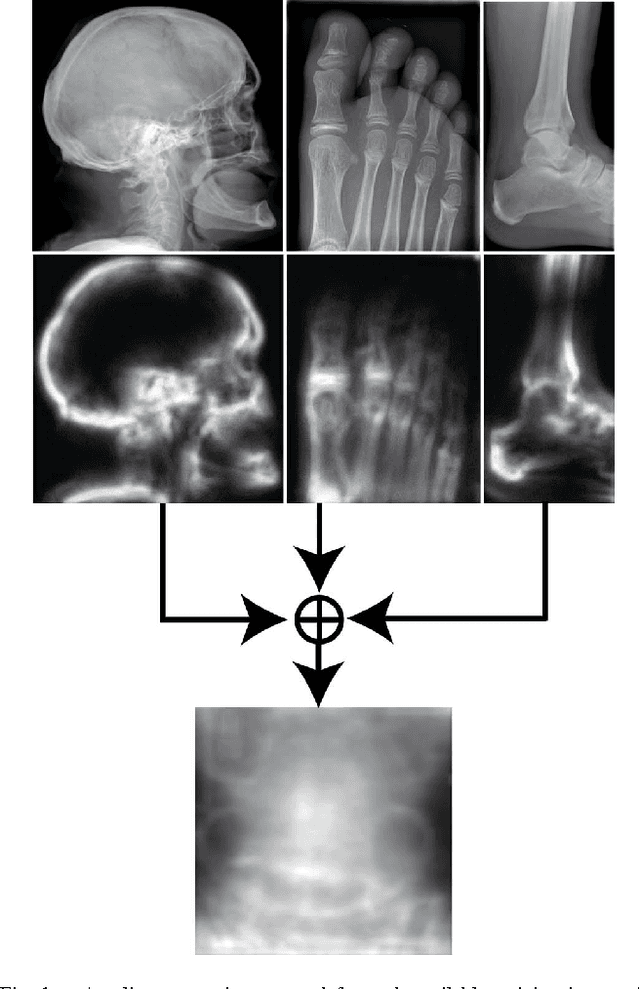
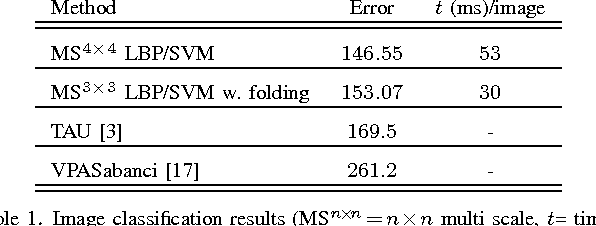
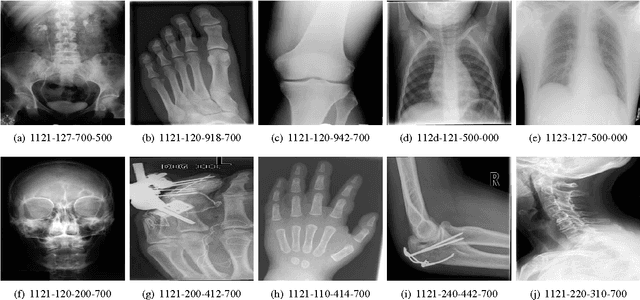
Abstract:Good results on image classification and retrieval using support vector machines (SVM) with local binary patterns (LBPs) as features have been extensively reported in the literature where an entire image is retrieved or classified. In contrast, in medical imaging, not all parts of the image may be equally significant or relevant to the image retrieval application at hand. For instance, in lung x-ray image, the lung region may contain a tumour, hence being highly significant whereas the surrounding area does not contain significant information from medical diagnosis perspective. In this paper, we propose to detect salient regions of images during training and fold the data to reduce the effect of irrelevant regions. As a result, smaller image areas will be used for LBP features calculation and consequently classification by SVM. We use IRMA 2009 dataset with 14,410 x-ray images to verify the performance of the proposed approach. The results demonstrate the benefits of saliency-based folding approach that delivers comparable classification accuracies with state-of-the-art but exhibits lower computational cost and storage requirements, factors highly important for big data analytics.
Autoencoding the Retrieval Relevance of Medical Images
Jul 05, 2015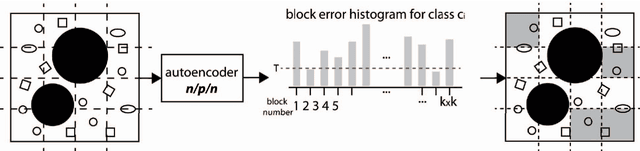
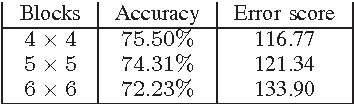
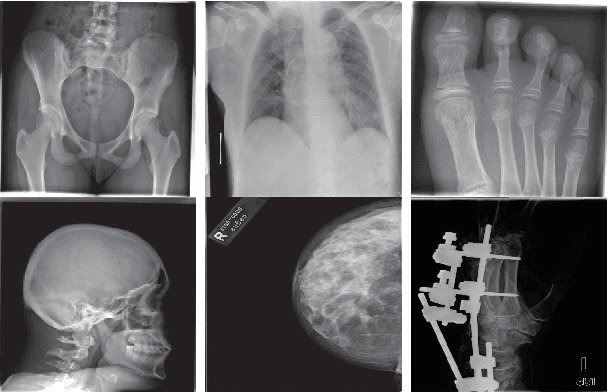
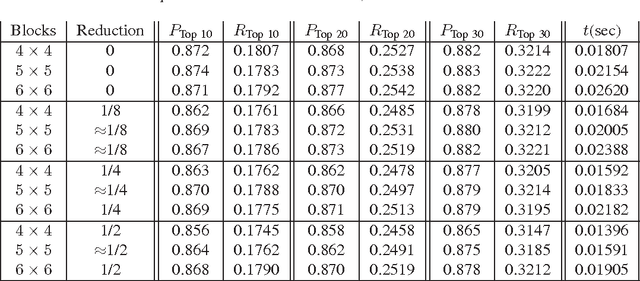
Abstract:Content-based image retrieval (CBIR) of medical images is a crucial task that can contribute to a more reliable diagnosis if applied to big data. Recent advances in feature extraction and classification have enormously improved CBIR results for digital images. However, considering the increasing accessibility of big data in medical imaging, we are still in need of reducing both memory requirements and computational expenses of image retrieval systems. This work proposes to exclude the features of image blocks that exhibit a low encoding error when learned by a $n/p/n$ autoencoder ($p\!<\!n$). We examine the histogram of autoendcoding errors of image blocks for each image class to facilitate the decision which image regions, or roughly what percentage of an image perhaps, shall be declared relevant for the retrieval task. This leads to reduction of feature dimensionality and speeds up the retrieval process. To validate the proposed scheme, we employ local binary patterns (LBP) and support vector machines (SVM) which are both well-established approaches in CBIR research community. As well, we use IRMA dataset with 14,410 x-ray images as test data. The results show that the dimensionality of annotated feature vectors can be reduced by up to 50% resulting in speedups greater than 27% at expense of less than 1% decrease in the accuracy of retrieval when validating the precision and recall of the top 20 hits.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge